Electronic Descriptor Based Qsar and Docking Studies of Some 2-[5-(Aryloxymethyl)-1, 3, 4-Oxadiazol-2-Ylsulfanyl] Acetic Acids as Anti-P.Aeruginosa
Adewole Oluwatosin Mary, Adeleke Babatunde Benjamin
Affiliation
1Department of Pure and Applied Chemistry, Ladoke Akintola, University of Technology, P.M.B. 4000, Ogbomoso, Oyo State, Nigeria
2Central Science Laboratory, Bowen University Iwo, Osun State, Nigeria
3Department of Chemistry, University of Ibadan, Ibadan, Nigeria
Corresponding Author
Adeleke Babatunde Benjamin, Department of Chemistry, University of Ibadan, Ibadan, Nigeria, Email: adelekebb46@gmail.com
Citation
Adeleke, B. B., et al. Electronic Descriptor Based Qsar and Docking Studies of Some 2-[5-(Aryloxymethyl) - 1, 3, 4-Oxadiazol-2-YIsulfanyl] Acetic Acids as Anti- P.Aeruginosa. (2018) J Pharm Pharmaceutics 5(2): 57- 60.
Copy rights
© 2018 Adeleke,B.B. This is an Open access article distributed under the terms of Creative Commons Attribution 4.0 International License.
Keywords
QSAR; DFT, 2-[5-(aryloxymethyl)-1, 3, 4-oxadiazol-2-ylsulfanyl] acetic acids; P.aeruginosa; Docking
Abstract
Molecular parameters of 2-[5-(aryloxymethyl)-1, 3, 4-oxadiazol-2-ylsulfanyl] acetic acids and it derivatives were calculated using density Functional theory (DFT). Quantitative Structure Activity Relationship (QSAR) was developed for the antimicrobial activity of the compounds while the interaction between the compounds and the receptor (4r0s) was studied using docking method. We report the molecular electronic descriptors for anti-P.aeruginosa activities of the seven compounds. Using the developed QSAR models, the predicted bioactivity (IC50) of the compounds fitted well with the experimentally observed IC50. In addition, ligand-receptor interactions are reported and 2-(5-((4-nitrophenoxy) methyl)-1, 3, 4-oxadiazol-2-ylsulfanyl) acetic acid (A7 ) showed the greatest affinity to bind on the active site of P. aeruginosa cell line
Introduction
Bacterial resistance to antibiotics remain a great threat to human health globally. Antibiotic resistance has been responsible for increasing bacterial infections among the young and the old[1]. There are still continuous great effort that are devoted to finding new and more efficient antibiotics that can supress the defence system of bacteria[2]. Therefore, it is in the search for new antimicrobial drug that can offer new pathway of action to solve the problem of multi-drug resistance that Neelam and co-workers (2016)[3] synthesized a number of 2-[5-(aryloxymethyl)-1, 3, 4-oxadiazol-2-ylsulfanyl] acetic acids with antibacterial properties against P.aeruginosa.
Pseudomonas aeruginosa is a bacterial pathogen that is metabolically resourceful. It can cause obstinate health issue in patients with blister and forfeited immune system[4].Pseudomonas aeruginosa also inhabits the surface of living things, such as animals, vegetation and human being[5]. It is a gram-negative bacteria and it is a cause of many infections like nosocomial infection, chronic airway infections etc[6]. It easily adapt to any change in location and only require few nutrients to grow and develop defence against antibiotics[7].
Furthermore, biological activities of compounds depend on their structure and electronic properties. Computational methods afford the opportunity to obtain information on the most stable conformation and electronic parameters of organic molecules. We therefore undertake in this work, computational investigation of the synthesized substituted acetic acids with a view to developing a quantitative structure activity relationship (QSAR) for these compounds and their antibacterial action on Pseudomonas aeruginosa.
Quantitative structural activity relationship (QSAR) as an arithmetic model is very useful in the prediction of the toxicity of material[8-10]. In QSAR study, descriptor selection is a vital step. A fundamental characteristic of building any QSAR model is the selection of suitable set of parameters with good predictability[11,12]. So, the use of quantum chemical parameters has remarkable importance[13,14].
In this work, the structures of 2-(5-(phenoxymethyl)-1,3,4-oxadiazol-2-ylsulfanyl) acetic acid (A1), 2-(5-((4- methylphenoxy) methyl)-1, 3, 4-oxadiazol-2-ylsulfanyl) acetic acid (A2), 2-(5-((4-methoxyphenoxy) methyl)-1, 3, 4-oxadiazol-2-ylsulfanyl) acetic acid (A3), 2-(5-((4-chlorophenoxy)methyl)-1,3,4-oxadiazol-2-ylsulfanyl) acetic acid (A4), 2-(5-((4-bromophenoxy)methyl)-1,3,4-oxadiazol-2-ylsulfanyl) acetic acid (A5), 2-(5-((4-fluorophenoxy)methyl)-1,3,4-oxadiazol-2-ylsulfanyl) acetic acid (A6) and2-(5-((4-nitrophenoxy)methyl)-1,3,4-oxadiazol-2-ylsulfanyl) acetic acid (A7) were optimised using density functional theory (DFT).Therefore, the molecular descriptors that describe the anti-P.aeruginosa activity of the studied compounds using DFT method were calculated and were used to develop QSAR model for examining the bioactivity of selected compounds. In addition, the optimized molecular compounds were docked against receptor (4r0s)[15] in order to obtain their binding energies with the receptor.
Computational details
Electronic descriptors and QSAR studies: Optimization of seven compounds was performed using 6-31(d,p) basis set so as to obtain molecular descriptors for anti-P.aeruginosa activity of 2-[5-(aryloxymethyl)-1, 3, 4-oxadiazol-2-ylsulfanyl] acetic acids and its analogues. The optimization was achieved using quantum chemical software i.e. Spartan ’14 by wave functionInc (Spartan 14)[16]. Also, QSAR model were developed using selected molecular descriptors in order to probe into biological activities of the studied compounds[17]. The development of the QSAR model was achieved by using multiple linear regression method and this was validated using cross validation (R2) and adjusted R2 (Equation 1 and 2).
------- (1)
The R2 adjusted could be calculated using equation (2)
------- (2)
Furthermore, docking study was performed for observing binding energy and non-bonding interaction between the optimized molecules and the P.aeruginosa cell line, MTCC 2453 receptor, (PDB ID: 4r0s). The software used for docking study were; Discovery Studio, Autodock tool, AutoDockvina and Edupymol version 1.7.4.4.
Result and Discussion
Molecular descriptors and QSAR studies: The optimized structures of the compounds are shown in Figure 1, while the calculated molecular descriptors are given in Table 1.
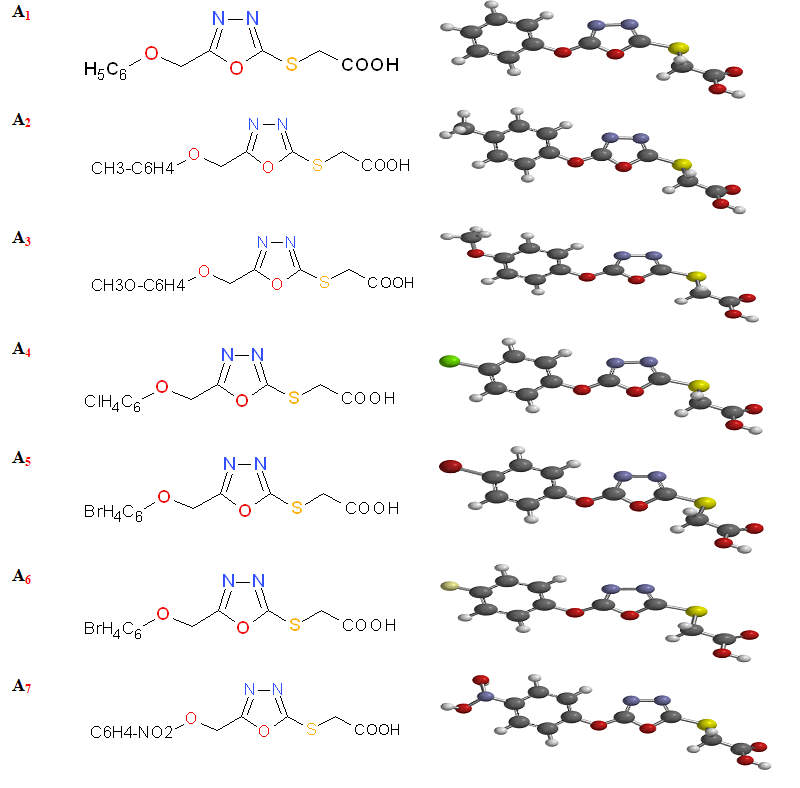 Figure 1: The schematic structures of the 2-[5-(aryloxymethyl)-1, 3, 4-oxadiazol-2-ylsulfanyl] acetic acids Derivatives.
Figure 1: The schematic structures of the 2-[5-(aryloxymethyl)-1, 3, 4-oxadiazol-2-ylsulfanyl] acetic acids Derivatives.
A QSAR investigation was executed to observe the structure activity relationship of seven 2-[5-(aryloxymethyl)-1, 3, 4-oxadiazol-2-ylsulfanyl] acetic acids derivatives with antibacterial properties. In QSAR studies, fitting and predictability are a function of the quality of a developed QSAR model[18]. Also, the development of QSAR model require dependent and independent variables via statistical analysis. In this work, the dependent variable was chosen to be P.aeruginosa cell line and the calculated parameters (Table 1) served as the independent variables. The multiple linear regression method was used to develop the model (equation 3) with selected descriptors.
IC50= 0.712470 -0.0187647(SE) -0.101480 (HBA) + 0.489555 (LUMO) ---- (3)
The result given in Table 2 and Table 3 show that the model reproduced the observed IC50 and this demonstrated the quality and the predicting power of the developed QSAR model. However, replicating power of the developed QSAR model, confirmed through their correlating coefficient (R2), is not enough to justify the actual efficacy of any QSAR model, hence, the stability of the model could be confirmed by considering cross-validation method and adjusted R2. The obtained CV. R2 as shown in Table 3 proved that the model is reliable and fitting since it is greater than the standard (0.5)[19,20]. Also, the value obtained for adjusted R2 was greater than the standard (0.6) which showed the effectiveness of the model. As shown in Table 2, the test model (equation 4) indicates that the calculated IC50 using the developed model is not considerably different from the observed IC50.
------ (4)
Table 1: The calculated molecular descriptors obtained for the studied compounds.
| HOMO | LUMO | BG | DM (Debye) | SE (Kj/mol) | LOGP | MW | OVALITY | PSA | HET | POL | P.aeruginosa(MTCC 2453) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | -6.46 | -0.85 | 5.61 | 4.54 | -48.49 | 3.20 | 252.25 | 1.41 | 68.15 | -1.054 | 58.09 | 0.60 |
| A2 | -6.30 | -0.82 | 5.48 | 4.67 | -47.41 | 3.69 | 266.27 | 1.44 | 68.19 | -1.054 | 59.60 | 0.60 |
| A3 | -5.94 | -0.78 | 5.16 | 3.90 | -52.66 | 3.07 | 282.27 | 1.46 | 75.14 | -1.058 | 60.40 | 0.60 |
| A4 | -6.52 | -0.99 | 5.53 | 4.71 | -46.76 | 3.76 | 286.69 | 1.44 | 68.00 | -1.059 | 59.22 | 0.50 |
| A5 | -6.52 | -1.03 | 5.49 | 4.65 | -48.21 | 4.03 | 331.14 | 1.44 | 68.07 | -1.055 | 59.59 | 0.50 |
| A6 | -6.44 | -0.9 | 5.54 | 4.57 | -42.20 | 3.36 | 270.24 | 1.42 | 68.04 | -1.061 | 58.48 | 0.45 |
| A7 | -6.14 | -0.89 | 5.25 | 4.68 | -55.09 | 2.84 | 299.26 | 1.48 | 116.63 | -1.056 | 60.43 | 0.40 |
Table 2: Predicted and observed IC50 values.
| Observed IC50 | Predicted IC50 | Deviation | Test Model |
|---|---|---|---|
| 0.60 | 0.60 | 0.00 | 0.00 |
| 0.60 | 0.59 | 0.01 | 0.41 |
| 0.60 | 0.61 | -0.01 | 0.41 |
| 0.50 | 0.50 | 0.00 | 0.00 |
| 0.50 | 0.50 | 0.00 | 0.00 |
| 0.45 | 0.45 | 0.00 | 0.00 |
| 0.40 | 0.40 | 0.00 | 0.00 |
Table 3: Statistical parameters for validation of QSAR model.
| N | p | R2 | CV.R2 | R2adj |
|---|---|---|---|---|
| 7 | 3 | 0.994 | 0.989 | 0.989 |
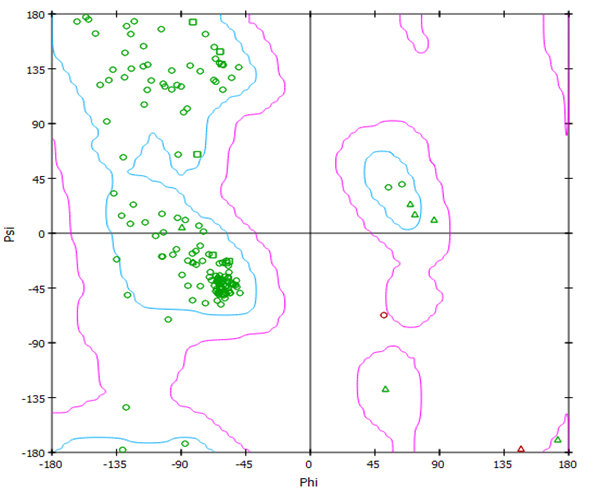
Docking studies: In this work, a modest assessment of the conformation of P.aeruginosa cell line (PDB ID: 4r0s) was observed using Ramachandran plot. As shown in Figure 2, the protein used in this work is stable and is of a good quality.
Figure 2: Ramachandran plot for P.aeruginosa cell line (PDB ID: 4r0s)
The seven molecular compounds were docked on the active gouge of P.aeruginosa cell line (PDB ID: 4r0s) and the outcome are shown on Table 4.
Table 4: Binding energy between ligands and 4r0s receptor.
| comp | Docking Binding Energy (kcal/mol) |
|---|---|
| A1 | -5.20 |
| A2 | -5.30 |
| A3 | -5.30 |
| A4 | -5.20 |
| A5 | -5.30 |
| A6 | -5.30 |
| A7 | -5.60 |
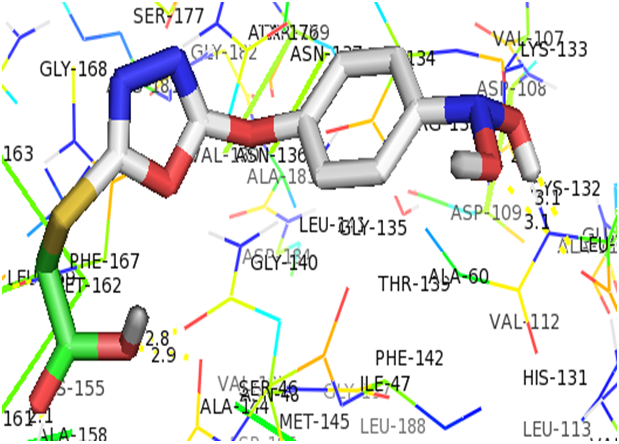
As displayed on Table 4, the binding energy for each complex were ranged from -5.20 to -5.60 kcal/mol and this revealed that A7 had the greatest affinity to bind to the active site of P.aeruginosa cell line with a binding energy of -5.60 kcal/mol. Nine conformations each were observed for the docking simulation of individual ligand-receptor complex and figure 3 shows the residue involved in the interaction between A7 and 4r0s. In the complex formed between A7 and the receptor, TYR-50, PRO-45 and ASN-48 formed coordination and hydrogen bond (with bond distance 2.1Å, 2.9Å and 2.8Å) with O<3, O4 and O4 of carboxylic acid of oxadiazole ring respectively. Also, LEU-61 formed hydrogen bond with O5 of nitro group of nitrobenzene twice and O6 of nitro group of nitrobenzene with bond distance 2.6Å, 3.1Å and 3.1Å respectively.
Figure 3: Interactions of A7 with the residue in the active gouge of P.aeruginosa cell line (4r0s).
Conclusion
Anti-P.aeruginosa activity of some 2-[5-(aryloxymethyl)-1, 3, 4-oxadiazol-2-ylsulfanyl] acetic acids was studied with the use of quantum chemical method via density functional theory to generate molecular descriptors for quantitative structural activity relationship study. The developed QSAR model replicated the observed bioactivities very well and this shows that the selected descriptors described the anti-P.aeruginosa properties of 2-[5-(aryloxymethyl)-1, 3, 4-oxadiazol-2-ylsulfanyl] acetic acid derivatives. More so, nine stable conformations were observed from docking study and 2-(5-((4-nitrophenoxy) methyl)-1, 3, 4-oxadiazol-2-ylsulfanyl) acetic acid inhibited P.aeruginosa cell line more than any other studied compounds.
References
- 1. Hancock, R.E.W., Sahl, H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. (2006) Nat Biotechnol 24: 1551-1557.
- 2. Christine, M.A., Ngoc, N.P., Christopher, A.A. Effect of Composition on Antibacterial Activity of Sequence-Defined Cationic Oligothioetheramides. (2018) ACS Infect Dis 4(8): 1257-1263.
- 3. Neelam, J., Deepika, S., Ruchika, G., et al. Synthesis and antibacterial screening of some 2-[5-(aryloxymethyl)-1, 3, 4-oxadiazol-2-ylsulfanyl] acetic acids. (2016) Der PharmaChemica 8(4): 434-438.
Pubmed|| Crossref|| Others
- 4. Ramos, J.L. (ed.): Pseudomonas. Heidelberg: Springer; (2004) : 1-7.
Pubmed|| Crossref|| Others
- 5. Klockgether, J., Tümmler, B. Recent advances in understanding Pseudomonas aeruginosaas a pathogen. (2017) F1000Res 6: 1261.
- 6. Meenakumari, S., Verma,S., Absar,A., et al. Antimicrobial susceptibility pattern of clinical isolates of P.aeruginosain an Indian cardiac Hospital. (2011) Internat J Engineer Sci Tech 3(9):7117-7124.
Pubmed|| Crossref|| Others
- 7. Strateva, T., Markova, B., Mitov, I., et al. Distribution of the type III effector proteins-encoding genes among nosocomial Pseudomonas aeruginosa isolates from Bulgaria. (2010) Ann Microbiol 60(3): 503-509.
Pubmed|| Crossref|| Others
- 8. Dahl, G.E., Jaitly, N., Salakhutdinov, R. Multitask Neural Networks for QSAR Predictions. (2014) arXiv preprint: 21.
Pubmed|| Crossref|| Others
- 9. Oyebamiji, A.K., Semire, B. DFT-QSAR model and docking studies of Antiliver cancer (HEPG-2) activities of 1, 4-diydropyridine based derivatives. (2016) Cancer Bio 6(2): 69-72.
Pubmed|| Crossref|| Others
- 10. Oyebamiji, A.K., Semire, B. Studies of antihypertensive activity of 1, 4dihydropyridine derivatives: combinations of DFT-QSAR and docking approaches. (2016) Bulletin Pharm Res 6(3): 105-118.
- 11. Guyon, I., Elisseeff, A. An introduction to variable and features selection. (2003) J. Mach. Learning Res. 3: 1157-1182.
Pubmed|| Crosssref|| Others
- 12. Nitendra, K.S., Mukesh, C.S., Vishnukanth, M., et al. QSAR studies of some side chain modified 7-chloro-4-aminoquinolines as antimalarial agents (2014). Arabian J Chem 7(5): 701-707.
- 13. Oyebamiji, K.A., Semire, B. Studies of 1, 4-dihydropyridine derivatives for anti-breast cancer (MCF-7) activities: combinations of DFT-QSAR and docking methods. (2016) New York Sci J 9(6): 58-66.
Pubmed|| Crossref|| Others
- 14. Zhu, M., Ge, F., Zhu, R., et al. A DFT-based QSAR study of the toxicity of quaternary ammonium compounds on Chlorella vulgaris. (2010) Chemosphere 80(1): 46-52.
- 15. Xu, K., Shanshan, L., Wen, Y., et al. Structural and Biochemical Analysis of Tyrosine Phosphatase Related to Biofilm Formation A (TpbA) from the Opportunistic Pathogen Pseudomonas aeruginosa PAO1. (2015) PLoS One 10(4): 0124330.
- 16. Spartan 14 wavefunction Inc. Irvine, CA 92612, USA.
Pubmed|| Crossref|| Others
- 17. Pourbasheer, E., Riahi, S., Ganjali, M.R., et al. Application of genetic algorithm support vector machine, (GA–SVM) for prediction of BK-channels activity. (2009) Eur J of Med Chem 44(12): 5023-5028.
- 18. Abolghasem, B., Eslam, P., Mehdi, N., et al. QSAR modeling of antimalarial activity of urea derivatives using genetic algorithm–multiple linear regressions. (2016) J Saudi Chem Soc 20(3): 282-290.
Pubmed|| Crossref|| Others
- 19. Marrero, P.Y., Torrens, F., Castro, E.A., et al. Atom, atom-type, and total linear indices of the molecular pseudograph’s atom adjacency matrix: application to QSPR/QSAR studies of organic compounds. (2004) Molecules 9(12): 1100-1123.
- 20. Oyebamiji, A.K., Semire, B. DFT-QSAR and Docking Studies of 2-[5-(aryloxymethyl)-1, 3, 4-oxadiazol-2-ylsulfanyl] acetic acids Derivatives against Bacillus subtilis. (2018) Der Pharma Chem 10(3): 135-139.
Pubmed|| Crossref|| Others